Why parallel
Humans are not designed to think parallel, instead we suffered from being interrupted and overberden by mutiple tasks. Modern computers usually implement multple CPUs, parallel/multiprocess concept was pretty common in programming languages.
In R, functions are nicely wrapped with minimum hardware details exposed, sometimes hidden functions has been parallel common algorithms at the backend. But how about these functions of your own creation?

One example with lapply
Think of the inverse sqrt which is a pretty common thing in distance calculation. Using lapply
trials <- runif(10000000,0,1)
rand <- runif(1000000000,0,1)
repeat_inverse_sqrt <- function(x)
{
arr <- runif(10000000,0,1)
return (1/sqrt(sum(arr^2)))
}
system.time(
lapply(trials, repeat_inverse_sqrt)
)
The total computing time as follows.
user system elapsed
98.135 1.552 100.635
Understanding the result:
user: time used by user process, in this case the RStudio main session.
system: time used system expense, system calls for example
elapsed: the actually time used.
We could see that basically elapsed = user + system, and user takes most of the time which make sense. The calculation takes around 100 seconds.
Using parallel in R
One of the nice packages for it will be “parallel”
Installing package
install.packages("parallel")
library(parallel)
using mclapply
system.time(
mclapply(trials, repeat_inverse_sqrt, mc.cores = cores)
)
user system elapsed
193.675 2.412 30.015
elapsed is down to 30 seconds!
– 1) Wait, what is happeding 193 seconds in user?
My little server has 8 cores. user means our process takes 193 seconds on 8 cores total.
– 2)Wait, why it is 193, while the simple one is 100
It is likely that 8 core of cpu is competing for memory resources since there is a lot data. Accessing GBs of data takes extra amount of time.
– 3) But why it is 30 not 193/8 = 25?
It is likely to be near that. while there still might be a sequential memory access for cpus and the process wait for every one to fininish.
|||||||||||||||||||||||||||||||||||| 0-24sec
|||||||||||||||||||||||||||||||||||| 1-25sec
|||||||||||||||||||||||||||||||||||| 2-26sec
|||||||||||||||||||||||||||||||||||| 3-27sec
|||||||||||||||||||||||||||||||||||| 7-31sec
Can we do better Parallel ?
Looking back to mclapply
The mechanism work behind mclapply is linux folk. You can google it, in short, it creates a all-mighty copy of master process and every single bit it uses to do things in parallel.
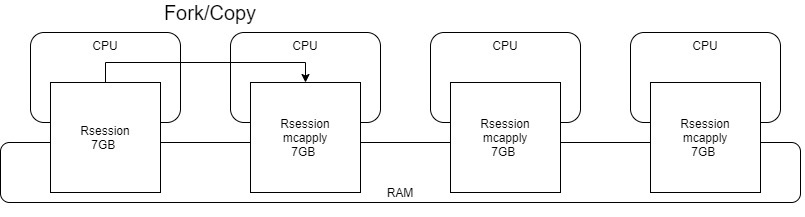
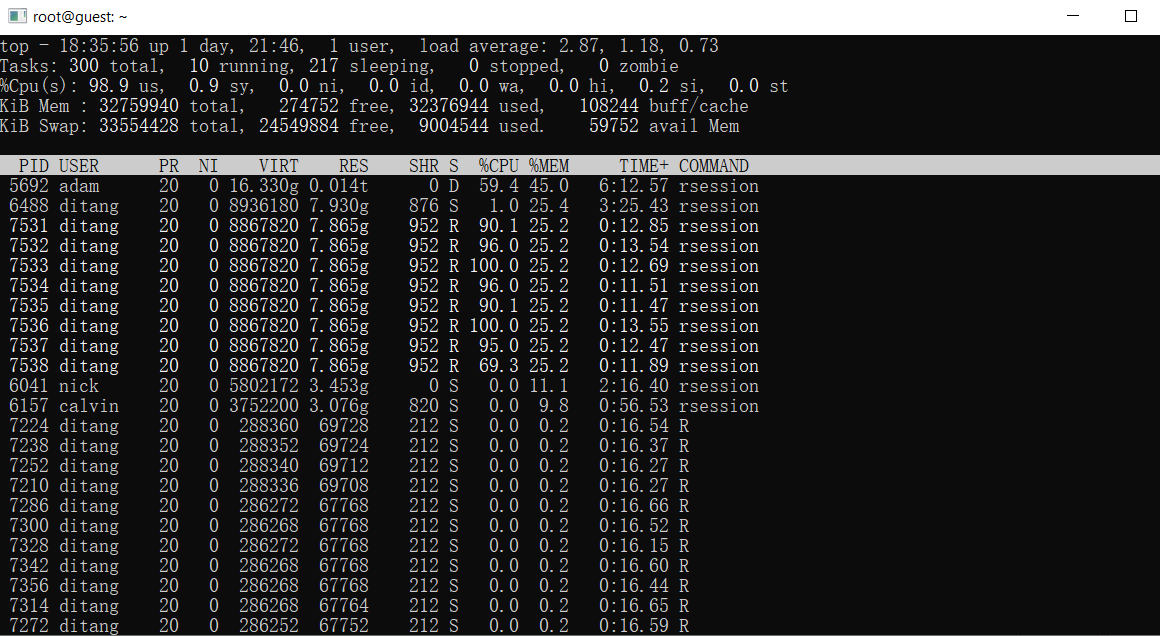
Resource usage in system. Every process is taking same amount of memory! That’s…. hmm. There are other variables in my working environment, garbages LOL.
Introducing parLapply
cl <- makeCluster(cores) # create a cluster
clusterExport(cl, "trials") # feed specific data to the clusters, excluding all others
system.time( # apply
parLapply(cl, trials, repeat_inverse_sqrt),
)
stopCluster(cl) # clean up messes
user system elapsed
1.077 0.216 27.744
elapsed is down to 27! And more importantly, it saves the ridiculous amount of memories.
why?
The reason was that parLapply creates minion processes that uses only what’s necessary and talk back to master through socket.
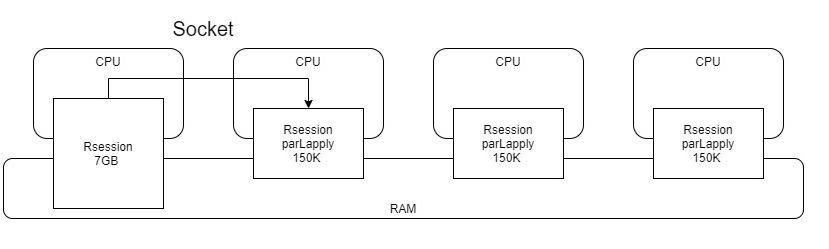
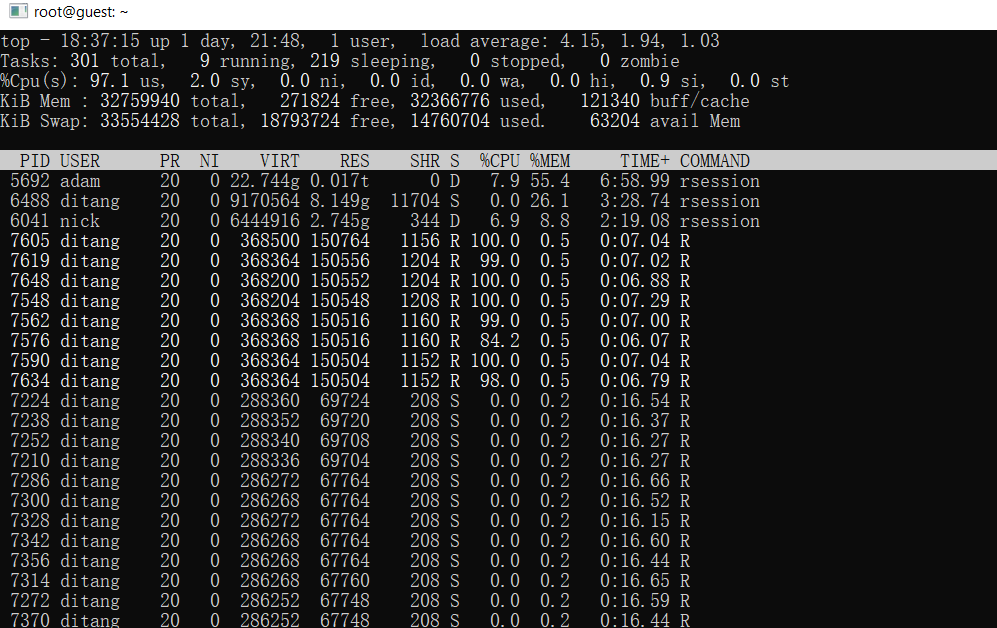
Resource usage in system. A lot less now! Depends on your project, this might be a your saver.
Summary
Although it is a little against instinct, having parallel thinking might be critical for improving performance your analytical programs.